Subproject 2: Transcriptomics / Proteomics
Spatio-temporal mapping of transcriptomic and proteomic heterogeneity of sarcomas
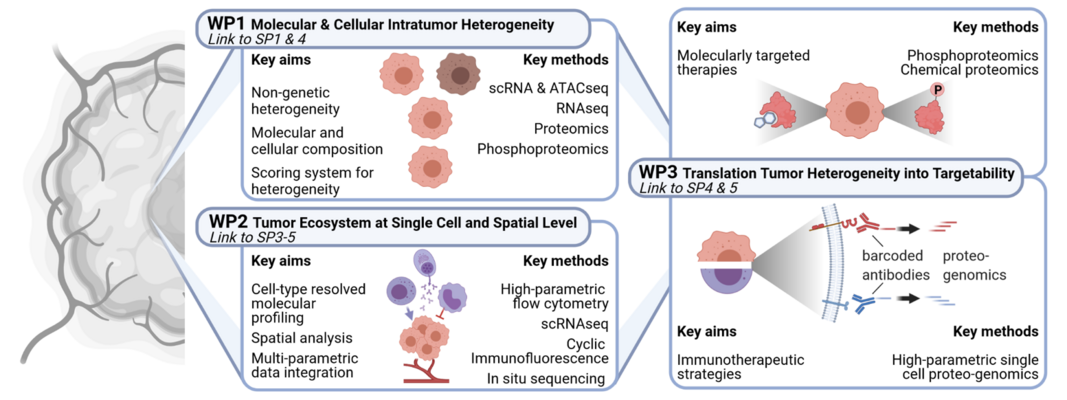
Neoplastic transformation in FDS is mainly driven by fusion genes encoding oncogenic transcription factors. Besides genomic aberrations, transcriptional plasticity and TME characterize the heterogeneity of FDS and determine disease progression, therapy resistance and metastatic potential. We will combine innovative and state-of-the-art single-cell and bulk technologies to systematically analyze the transcriptomic and proteomic features of FDS ecosystems to (i) decipher ITH at the level of transcripts, proteins and cell types; (ii) understand the role of the TME in tumor immune evasion, therapy resistance and metastasis, and (iii) translate insights into ITH into potentially feasible therapeutic strategies. These datasets will be integrated with those from subprojects 1 and 3 in subproject 4 and will serve as an essential resource for subproject 5, which will functionally validate the identified resistance mechanisms and explore novel therapeutic targets.
Main objectives:
-
Deciphering the cellular sarcoma ecosystem at the single cell level
-
Temporal mapping of transcriptomic and proteomic heterogeneity of sarcomas
-
Spatial mapping of transcriptomic and proteomic heterogeneity of sarcomas